Research
Overview: Nucleic Acid Chemistry and Engineering
 Nucleic acids have been chosen as the chemical building blocks in the evolution of life. Inspired by Nature, DNA/RNA nanotechnology emerges to allow precise spatial organization and dynamic control of information networks. The programmability of sequence-specific self-assembly and innate biocompatibility have made it possible for these unique nanostructures and nanodevices to interact in biological matrices. Our research group is interested in creating synthetic DNA- and RNA-based tools for analytical and biomedical applications. Our research program integrates the disciplines of chemistry, bionanotechnology, cellular biology, and synthetic biology. Our ultimate goal is to understand the fundamentals of life, and to construct artificial genetic devices and pathways.
Nucleic acids have been chosen as the chemical building blocks in the evolution of life. Inspired by Nature, DNA/RNA nanotechnology emerges to allow precise spatial organization and dynamic control of information networks. The programmability of sequence-specific self-assembly and innate biocompatibility have made it possible for these unique nanostructures and nanodevices to interact in biological matrices. Our research group is interested in creating synthetic DNA- and RNA-based tools for analytical and biomedical applications. Our research program integrates the disciplines of chemistry, bionanotechnology, cellular biology, and synthetic biology. Our ultimate goal is to understand the fundamentals of life, and to construct artificial genetic devices and pathways.
Primary Research Interests
(1) Biosensors and Bioimaging:
 Microscopy becomes a central tool to study the spatial and temporal dynamics of cellular components. Our research focus is to engineer imaging probes to quantify and track the synthesis, function, and degradation pathways of metabolites, signaling molecules, antibiotics, and other small molecules in live cells. These imaging probes will be constructed based on our knowledge about conditional self-assembly of nucleic acid nanostructures and integrated circuits.
Microscopy becomes a central tool to study the spatial and temporal dynamics of cellular components. Our research focus is to engineer imaging probes to quantify and track the synthesis, function, and degradation pathways of metabolites, signaling molecules, antibiotics, and other small molecules in live cells. These imaging probes will be constructed based on our knowledge about conditional self-assembly of nucleic acid nanostructures and integrated circuits.
Ren (2020) JACS, 142, 2968-2974; Rigumula (2019) Angew Chem Int Ed, 58, 18271-18275; You (2019) Cell Chem Biol, 26, 471-481; Karunanayake (2018) JACS, 140, 8739-8745; You (2015) PNAS, 112, E2756-E2765.
(2) Cell Membrane Biophysics:
We recently developed an efficient lipid-based approach to immobilize designer DNA probes outside of live cell membranes. With this approach, we are developing novel DNA probes that can measure transient membrane lipid encounter events and other biophysical events at cell membranes, such as cellular mechanotransduction.
Tian (2021) Chem Sci, 12, in press; Zhao (2020) Chem Sci, 11, 8558-8566; Bagheri (2019) Chem Sci, 10, 11030-11040; Zhao (2017) JACS, 139, 18182-18185; You (2017) Nat Nanotechnol, 12, 453-459;
(3) DNA/RNA Nanotechnology:
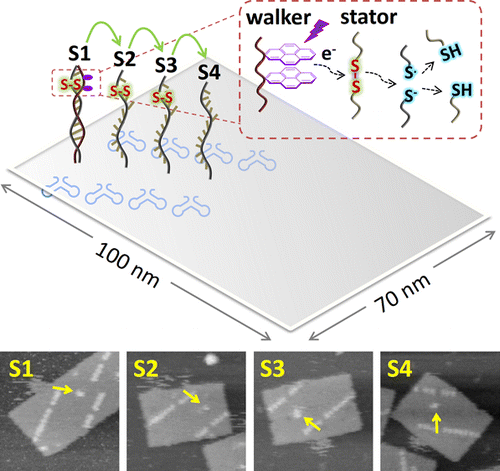
DNA/RNA nanotechnology has succeeded in constructing precisely-defined nanostructures and nanodevices in vitro. Nucleic acid tools are great choice to study biology, especially considering their programmability of sequence-specific self-assembly and innate biocompatibility. Our research focuses on the construction of genetically encoded RNA nanostructures in live cells. We are interested in discovering novel molecular mechanisms for precise conditional self-assembly of these nucleic acid nanostructures.
Han#, Wu#, You# (2015) Nat Chem, 7, 835-841; You (2015) JACS, 137, 667-674; You (2014) JACS, 136, 1256-1259; You (2012) ACS Nano, 6, 7935-7941.
(4) SELEX:
Aptamer is single-stranded DNA or RNA that can specifically recognize a target of interest. Systematic evolution of ligands by exponential enrichment (SELEX) is a high-throughput technique to generate these functional nucleic acids. Our research interest is to identify aptamers and catalytic RNAs that function inside live cells. These identified RNAs or DNAs will be further applied to construct artificial biological devices and pathways that does not exist in Nature.
You (2011) Chem Sci, 2, 1003-1010; Liu#, You# (2011) Curr Med Chem, 18, 4117-4125.
(5) Photo-controlled Devices:
 Light can be used to externally manipulate the functions of nanodevices with both spatial and temporal precision. The use of light to control intracellular structure and function of proteins is generally termed optogenetics. We are interested in developing optogenetically controlled RNA devices by taking advantage of photoswitchable chromophores.
Light can be used to externally manipulate the functions of nanodevices with both spatial and temporal precision. The use of light to control intracellular structure and function of proteins is generally termed optogenetics. We are interested in developing optogenetically controlled RNA devices by taking advantage of photoswitchable chromophores.
Ren (2020) Angew Chem Int Ed, 59, 218986-21990; You (2015) Ann NY Acad Sci, 1352, 13-19; You (2012) Angew Chem Int Ed, 51, 2457-2460.